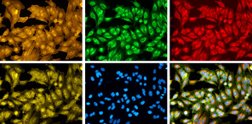
The public-private translational center Ksilink together with industry and non-profit partners recently joined a collaborative initiative, launched and coordinated by the Broad Institute of MIT and Harvard, in orderto create a large cell-imaging dataset, displaying more than 1 billion cells responding to over 140,000 small molecules and genetic perturbations.
This microscopy image dataset will represent the largest collection of cell images generated by Cell Painting and will act as a reference collection to potentially fuel efforts for discovering and developing new therapeutics. Twelve very renowned partners from the biotechnology and pharmaceutical industries have joined Broad Institute’s effort, called the Joint Undertaking in Morphological Profiling with Cell Painting (JUMP-CP).
Once the image dataset is created, it will be available to consortium members for the first year, after which it will be freely available for use by the scientific community. With the complete reference dataset, potential applications include predicting a compounds activity and toxicity in cells, matching appropriate drugs to different disease states, and more.
Powering patient-based, phenotypic drug discovery and development with images
Ksilink aims in identifying and developing new drugs by using cellular models directly developed from cellular material from patients with genetic mutations. These physiologically highly relevant disease models are used for phenotypic high-throughput high content screening. Cell microscopy images contain a vast amount of information about the status of the cell: whether it is diseased, how it is responding to a drug treatment, or whether a certain pathway has been disrupted, for example.
In this context Ksilink develops image data analysis (or computer vision) techniques for analyzing multi-parametric cellular patterns with the use of deep-learning algorithms. The use of Artificial Intelligence makes it possible to detect even very subtle structural changes in the cells when they are exposed to a potentially active compound.
This approach is described as target agnostic: Ksilink does not focus on a specific target molecule or signal transduction pathway, but instead tests the compounds in the complete, complex cell system – which is called phenotypic or cell-based screening.
The cell imaging analysis, as undertaken in the framework of the JUMP-CP Project, can determine the underlying mechanisms at work when cells are affected by chemical compounds. It allows Ksilink to get access to an annotated set of over 140,000 structures from small molecules and genetic perturbations and thus can foster the determination of the mechanisms of action of active compounds identified in phenotypic screening campaigns.
The JUMP-CP team, coordinated at Broad Institute by Dr Anne Carpenter, will create a critical mass of cellular imaging data to empower discoveries about cell biology that can inform drug discovery and development. The goal is to identify the effect of each gene or compound on the cell’s shape or activity — creating a morphological atlas that researchers can reference as a baseline in their own studies. With a large reference of image-based cellular profiles, scientists could computationally compare their images to determine a drug’s likely mechanism of action or a gene variant’s impact, prior to any clinical trials in patients.
“Ultimately, we aim to make cell images as accessible and analyzable as genomes and transcriptomes,” says Carpenter.
“Ksilink is very happy to make part of this prestigious consortium. It allows us to improve our capacities to identify new classes of molecules for the development of drug candidates for diseases with great medical needs” says De Lacombe, Managing Director of Ksilink.
Building an exquisite reference dataset, based on Broad Institute’s knowledge
The consortium will use the Broad Institute's flagship assay, Cell Painting, to systematically survey cell morphology and build the reference dataset. Metrics extracted from cellular images will capture information on dozens of features to mark how the cell was affected by each chemical compound or genetic perturbation, allowing scientists to explore the underlying biology with the power of automated image analysis. The collaboration is funded in part by the Bits to Bytes Capital Call of the Massachusetts Life Sciences Center (MLSC).
For more information, please also see the reference article, published by Broad Institute: https://www.broadinstitute.org/news/broad-institute-launches-academic-industry-cell-imaging-consortium-speed-drug-discovery-and